In this video, I will be showing you how to easily web scrape data from websites in Python using the pandas library. Particularly, the readhtml function o. Web Scraping with Pandas and Beautifulsoup Web scraping. Pandas has a neat concept known as a DataFrame. A DataFrame can hold data and be easily manipulated. Converting to lists. Rows can be converted to Python lists. Pretty print pandas dataframe. You can convert it to an ascii table with the.
How to Setup the Scraping Project. Our setup is pretty simple. Just create a folder and install Beautiful Soup, pandas, and requests. To create a folder and install the libraries, enter the commands given below. I am assuming that you have already installed Python 3.x. Mkdir scraper pip install beautifulsoup4 pip install requests pip install pandas. How to Scrape a Website with a Single Line of Python Code Pandas Makes Getting Table Data Easy. I've read lengthy tutorials where the authors have used libraries like urllib and BeautifulSoup and many, many steps to scrape and parse sports statistics from web pages. The pandas.readhtml function uses some scraping libraries such as BeautifulSoup and Urllib to return a list containing all the tables in a page as DataFrames. You just need to pass the URL of the page.
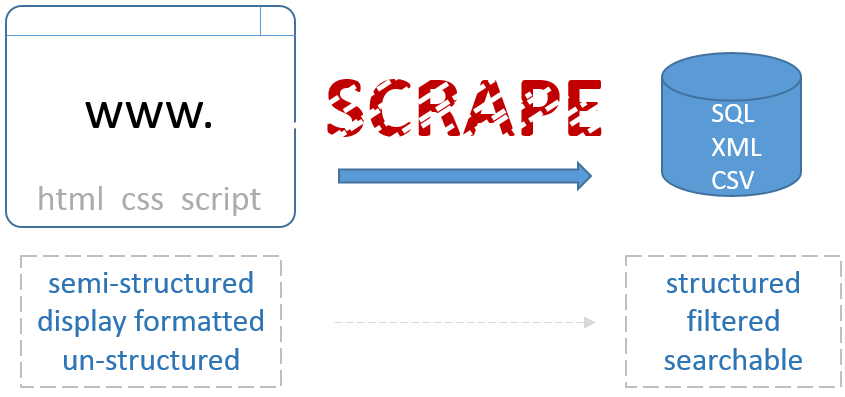
Part one of this series focuses on requesting and wrangling HTML using two of the most popular Python libraries for web scraping: requests and BeautifulSoup
After the 2016 election I became much more interested in media bias and the manipulation of individuals through advertising. This series will be a walkthrough of a web scraping project that monitors political news from both left and right wing media outlets and performs an analysis on the rhetoric being used, the ads being displayed, and the sentiment of certain topics.
The first part of the series will we be getting media bias data and focus on only working locally on your computer, but if you wish to learn how to deploy something like this into production, feel free to leave a comment and let me know.
You should already know:
- Python fundamentals - lists, dicts, functions, loops - learn on Coursera
- Basic HTML
You will have learned:
- Requesting web pages
- Parsing HTML
- Saving and loading scraped data
- Scraping multiple pages in a row
Every time you load a web page you're making a request to a server, and when you're just a human with a browser there's not a lot of damage you can do. With a Python script that can execute thousands of requests a second if coded incorrectly, you could end up costing the website owner a lot of money and possibly bring down their site (see Denial-of-service attack (DoS)).
With this in mind, we want to be very careful with how we program scrapers to avoid crashing sites and causing damage. Every time we scrape a website we want to attempt to make only one request per page. We don't want to be making a request every time our parsing or other logic doesn't work out, so we need to parse only after we've saved the page locally.
If I'm just doing some quick tests, I'll usually start out in a Jupyter notebook because you can request a web page in one cell and have that web page available to every cell below it without making a new request. Since this article is available as a Jupyter notebook, you will see how it works if you choose that format.
After we make a request and retrieve a web page's content, we can store that content locally with Python's open() function. To do so we need to use the argument wb, which stands for 'write bytes'. This let's us avoid any encoding issues when saving.
Below is a function that wraps the open() function to reduce a lot of repetitive coding later on:
Assume we have captured the HTML from google.com in html, which you'll see later how to do. After running this function we will now have a file in the same directory as this notebook called google_com that contains the HTML.
To retrieve our saved file we'll make another function to wrap reading the HTML back into html. We need to use rb for 'read bytes' in this case.
The open function is doing just the opposite: read the HTML from google_com. If our script fails, notebook closes, computer shuts down, etc., we no longer need to request Google again, lessening our impact on their servers. While it doesn't matter much with Google since they have a lot of resources, smaller sites with smaller servers will benefit from this.
I save almost every page and parse later when web scraping as a safety precaution.
Each site usually has a robots.txt on the root of their domain. This is where the website owner explicitly states what bots are allowed to do on their site. Simply go to example.com/robots.txt and you should find a text file that looks something like this:
The User-agent field is the name of the bot and the rules that follow are what the bot should follow. Some robots.txt will have many User-agents with different rules. Common bots are googlebot, bingbot, and applebot, all of which you can probably guess the purpose and origin of.
We don't really need to provide a User-agent when scraping, so User-agent: * is what we would follow. A * means that the following rules apply to all bots (that's us).
The Crawl-delay tells us the number of seconds to wait before requests, so in this example we need to wait 10 seconds before making another request.
Allow gives us specific URLs we're allowed to request with bots, and vice versa for Disallow. In this example we're allowed to request anything in the /pages/subfolder which means anything that starts with example.com/pages/. On the other hand, we are disallowed from scraping anything from the /scripts/subfolder.
Many times you'll see a * next to Allow or Disallow which means you are either allowed or not allowed to scrape everything on the site.
Sometimes there will be a disallow all pages followed by allowed pages like this:
This means that you're not allowed to scrape anything except the subfolder /pages/. Essentially, you just want to read the rules in order where the next rule overrides the previous rule.
This project will primarily be run through a Jupyter notebook, which is done for teaching purposes and is not the usual way scrapers are programmed. After showing you the pieces, we'll put it all together into a Python script that can be run from command line or your IDE of choice.
With Python's requests (pip install requests) library we're getting a web page by using get() on the URL. The response r contains many things, but using r.content will give us the HTML. Once we have the HTML we can then parse it for the data we're interested in analyzing.
There's an interesting website called AllSides that has a media bias rating table where users can agree or disagree with the rating.
Since there's nothing in their robots.txt that disallows us from scraping this section of the site, I'm assuming it's okay to go ahead and extract this data for our project. Let's request the this first page:
Since we essentially have a giant string of HTML, we can print a slice of 100 characters to confirm we have the source of the page. Let's start extracting data.
What does BeautifulSoup do?
We used requests to get the page from the AllSides server, but now we need the BeautifulSoup library (pip install beautifulsoup4) to parse HTML and XML. When we pass our HTML to the BeautifulSoup constructor we get an object in return that we can then navigate like the original tree structure of the DOM.
This way we can find elements using names of tags, classes, IDs, and through relationships to other elements, like getting the children and siblings of elements.
We create a new BeautifulSoup object by passing the constructor our newly acquired HTML content and the type of parser we want to use:
This soup object defines a bunch of methods — many of which can achieve the same result — that we can use to extract data from the HTML. Let's start with finding elements.
To find elements and data inside our HTML we'll be using select_one, which returns a single element, and select, which returns a list of elements (even if only one item exists). Both of these methods use CSS selectors to find elements, so if you're rusty on how CSS selectors work here's a quick refresher:
A CSS selector refresher
- To get a tag, such as
<a></a>,<body></body>, use the naked name for the tag. E.g.select_one('a')gets an anchor/link element,select_one('body')gets the body element .tempgets an element with a class of temp, E.g. to get<a></a>useselect_one('.temp')#tempgets an element with an id of temp, E.g. to get<a></a>useselect_one('#temp').temp.examplegets an element with both classes temp and example, E.g. to get<a></a>useselect_one('.temp.example').temp agets an anchor element nested inside of a parent element with class temp, E.g. to get<div><a></a></div>useselect_one('.temp a'). Note the space between.tempanda..temp .examplegets an element with class example nested inside of a parent element with class temp, E.g. to get<div><a></a></div>useselect_one('.temp .example'). Again, note the space between.tempand.example. The space tells the selector that the class after the space is a child of the class before the space.- ids, such as
<a id=one></a>, are unique so you can usually use the id selector by itself to get the right element. No need to do nested selectors when using ids.
There's many more selectors for for doing various tasks, like selecting certain child elements, specific links, etc., that you can look up when needed. The selectors above get us pretty close to everything we would need for now.
Tips on figuring out how to select certain elements
Most browsers have a quick way of finding the selector for an element using their developer tools. In Chrome, we can quickly find selectors for elements by
- Right-click on the the element then select 'Inspect' in the menu. Developer tools opens and and highlights the element we right-clicked
- Right-click the code element in developer tools, hover over 'Copy' in the menu, then click 'Copy selector'
Sometimes it'll be a little off and we need to scan up a few elements to find the right one. Here's what it looks like to find the selector and Xpath, another type of selector, in Chrome:
Our data is housed in a table on AllSides, and by inspecting the header element we can find the code that renders the table and rows. What we need to do is select all the rows from the table and then parse out the information from each row.
Here's how to quickly find the table in the source code:
Simplifying the table's HTML, the structure looks like this (comments <!-- --> added by me):
So to get each row, we just select all <tr> inside <tbody>:

tbody tr tells the selector to extract all <tr> (table row) tags that are children of the <tbody> body tag. If there were more than one table on this page we would have to make a more specific selector, but since this is the only table, we're good to go.
Now we have a list of HTML table rows that each contain four cells:
- News source name and link
- Bias data
- Agreement buttons
- Community feedback data
Below is a breakdown of how to extract each one.
The outlet name (ABC News) is the text of an anchor tag that's nested inside a <td> tag, which is a cell — or table data tag.

Getting the outlet name is pretty easy: just get the first row in rows and run a select_one off that object:
The only class we needed to use in this case was .source-title since .views-field looks to be just a class each row is given for styling and doesn't provide any uniqueness.
Notice that we didn't need to worry about selecting the anchor tag a that contains the text. When we use .text is gets all text in that element, and since 'ABC News' is the only text, that's all we need to do. Bear in mind that using select or select_one will give you the whole element with the tags included, so we need .text to give us the text between the tags.
.strip() ensures all the whitespace surrounding the name is removed. Many websites use whitespace as a way to visually pad the text inside elements so using strip() is always a good idea.
You'll notice that we can run BeautifulSoup methods right off one of the rows. That's because the rows become their own BeautifulSoup objects when we make a select from another BeautifulSoup object. On the other hand, our name variable is no longer a BeautifulSoup object because we called .text.
We also need the link to this news source's page on AllSides. If we look back at the HTML we'll see that in this case we do want to select the anchor in order to get the href that contains the link, so let's do that:
It is a relative path in the HTML, so we prepend the site's URL to make it a link we can request later.
Getting the link was a bit different than just selecting an element. We had to access an attribute (href) of the element, which is done using brackets, like how we would access a Python dictionary. This will be the same for other attributes of elements, like src in images and videos.
We can see that the rating is displayed as an image so how can we get the rating in words? Looking at the HTML notice the link that surrounds the image has the text we need:
We could also pull the alt attribute, but the link looks easier. Let's grab it:
Here we selected the anchor tag by using the class name and tag together: .views-field-field-bias-image is the class of the <td> and <a> is for the anchor nested inside.
After that we extract the href just like before, but now we only want the last part of the URL for the name of the bias so we split on slashes and get the last element of that split (left-center).
The last thing to scrape is the agree/disagree ratio from the community feedback area. The HTML of this cell is pretty convoluted due to the styling, but here's the basic structure:
The numbers we want are located in two span elements in the last div. Both span elements have classes that are unique in this cell so we can use them to make the selection:
Using .text will return a string, so we need to convert them to integers in order to calculate the ratio.
Side note: If you've never seen this way of formatting print statements in Python, the f at the front allows us to insert variables right into the string using curly braces. The :.2f is a way to format floats to only show two decimals places.
If you look at the page in your browser you'll notice that they say how much the community is in agreement by using 'somewhat agree', 'strongly agree', etc. so how do we get that? If we try to select it:
It shows up as None because this element is rendered with Javascript and requests can't pull HTML rendered with Javascript. We'll be looking at how to get data rendered with JS in a later article, but since this is the only piece of information that's rendered this way we can manually recreate the text.
To find the JS files they're using, just CTRL+F for '.js' in the page source and open the files in a new tab to look for that logic.
It turned out the logic was located in the eleventh JS file and they have a function that calculates the text and color with these parameters:
| Range | Agreeance |
| $ratio > 3$ | absolutely agrees |
| $2 < ratio leq 3$ | strongly agrees |
| $1.5 < ratio leq 2$ | agrees |
| $1 < ratio leq 1.5$ | somewhat agrees |
| $ratio = 1$ | neutral |
| $0.67 < ratio < 1$ | somewhat disgrees |
| $0.5 < ratio leq 0.67$ | disgrees |
| $0.33 < ratio leq 0.5$ | strongly disagrees |
| $ratio leq 0.33$ | absolutely disagrees |
Now that we have the general logic for a single row and we can generate the agreeance text, let's create a loop that gets data from every row on the first page:
In the loop we can combine any multi-step extractions into one to create the values in the least number of steps.
Our data list now contains a dictionary containing key information for every row.
Keep in mind that this is still only the first page. The list on AllSides is three pages long as of this writing, so we need to modify this loop to get the other pages.
Notice that the URLs for each page follow a pattern. The first page has no parameters on the URL, but the next pages do; specifically they attach a ?page=#to the URL where '#' is the page number.
Right now, the easiest way to get all pages is just to manually make a list of these three pages and loop over them. If we were working on a project with thousands of pages we might build a more automated way of constructing/finding the next URLs, but for now this works.
According to AllSides' robots.txt we need to make sure we wait ten seconds before each request.
Our loop will:
- request a page
- parse the page
- wait ten seconds
- repeat for next page.
Remember, we've already tested our parsing above on a page that was cached locally so we know it works. You'll want to make sure to do this before making a loop that performs requests to prevent having to reloop if you forgot to parse something.
By combining all the steps we've done up to this point and adding a loop over pages, here's how it looks:
Now we have a list of dictionaries for each row on all three pages.
To cap it off, we want to get the real URL to the news source, not just the link to their presence on AllSides. To do this, we will need to get the AllSides page and look for the link.
If we go to ABC News' page there's a row of external links to Facebook, Twitter, Wikipedia, and the ABC News website. The HTML for that sections looks like this:
Notice the anchor tag (<a>) that contains the link to ABC News has a class of 'www'. Pretty easy to get with what we've already learned:
So let's make another loop to request the AllSides page and get links for each news source. Unfortunately, some pages don't have a link in this grey bar to the news source, which brings up a good point: always account for elements to randomly not exist.
Up until now we've assumed elements exist in the tables we scraped, but it's always a good idea to program scrapers in way so they don't break when an element goes missing.
Using select_one or select will always return None or an empty list if nothing is found, so in this loop we'll check if we found the website element or not so it doesn't throw an Exception when trying to access the href attribute.
Finally, since there's 265 news source pages and the wait time between pages is 10 seconds, it's going to take ~44 minutes to do this. Instead of blindly not knowing our progress, let's use the tqdm library (pip install tqdm) to give us a nice progress bar:
tqdm is a little weird at first, but essentially tqdm_notebook is just wrapping around our data list to produce a progress bar. We are still able to access each dictionary, d, just as we would normally. Note that tqdm_notebook is only for Jupyter notebooks. In regular editors you'll just import tqdm from tqdm and use tqdm instead.
So what do we have now? At this moment, data is a list of dictionaries, each of which contains all the data from the tables as well as the websites from each individual news source's page on AllSides.
The first thing we'll want to do now is save that data to a file so we don't have to make those requests again. We'll be storing the data as JSON since it's already in that form anyway:
If you're not familiar with JSON, just quickly open allsides.json in an editor and see what it looks like. It should look almost exactly like what data looks like if we print it in Python: a list of dictionaries.
Before ending this article I think it would be worthwhile to actually see what's interesting about this data we just retrieved. So, let's answer a couple of questions.
Which ratings for outlets does the communityabsolutely agreeon?
To find where the community absolutely agrees we can do a simple list comprehension that checks each dict for the agreeance text we want:
Using some string formatting we can make it look somewhat tabular. Interestingly, C-SPAN is the only center bias that the community absolutely agrees on. The others for left and right aren't that surprising.
Which ratings for outlets does the communityabsolutely disagreeon?
To make analysis a little easier, we can also load our JSON data into a Pandas DataFrame as well. This is easy with Pandas since they have a simple function for reading JSON into a DataFrame.

As an aside, if you've never used Pandas (pip install pandas), Matplotlib (pip install matplotlib), or any of the other data science libraries, I would definitely recommend checking out Jose Portilla's data science course for a great intro to these tools and many machine learning concepts.
Now to the DataFrame:
| agree | agree_ratio | agreeance_text | allsides_page | bias | disagree | |
|---|---|---|---|---|---|---|
| name | ||||||
| ABC News | 8355 | 1.260371 | somewhat agrees | https://www.allsides.com/news-source/abc-news-... | left-center | 6629 |
| Al Jazeera | 1996 | 0.694986 | somewhat disagrees | https://www.allsides.com/news-source/al-jazeer... | center | 2872 |
| AllSides | 2615 | 2.485741 | strongly agrees | https://www.allsides.com/news-source/allsides-0 | allsides | 1052 |
| AllSides Community | 1760 | 1.668246 | agrees | https://www.allsides.com/news-source/allsides-... | allsides | 1055 |
| AlterNet | 1226 | 2.181495 | strongly agrees | https://www.allsides.com/news-source/alternet | left | 562 |
| agree | agree_ratio | agreeance_text | allsides_page | bias | disagree | |
|---|---|---|---|---|---|---|
| name | ||||||
| CNBC | 1239 | 0.398905 | strongly disagrees | https://www.allsides.com/news-source/cnbc | center | 3106 |
| Quillette | 45 | 0.416667 | strongly disagrees | https://www.allsides.com/news-source/quillette... | right-center | 108 |
| The Courier-Journal | 64 | 0.410256 | strongly disagrees | https://www.allsides.com/news-source/courier-j... | left-center | 156 |
| The Economist | 779 | 0.485964 | strongly disagrees | https://www.allsides.com/news-source/economist | left-center | 1603 |
| The Observer (New York) | 123 | 0.484252 | strongly disagrees | https://www.allsides.com/news-source/observer | center | 254 |
| The Oracle | 33 | 0.485294 | strongly disagrees | https://www.allsides.com/news-source/oracle | center | 68 |
| The Republican | 108 | 0.392727 | strongly disagrees | https://www.allsides.com/news-source/republican | center | 275 |
It looks like much of the community disagrees strongly with certain outlets being rated with a 'center' bias.
Let's make a quick visualization of agreeance. Since there's too many news sources to plot so let's pull only those with the most votes. To do that, we can make a new column that counts the total votes and then sort by that value:
| agree | agree_ratio | agreeance_text | allsides_page | bias | disagree | total_votes | |
|---|---|---|---|---|---|---|---|
| name | |||||||
| CNN (Web News) | 22907 | 0.970553 | somewhat disagrees | https://www.allsides.com/news-source/cnn-media... | left-center | 23602 | 46509 |
| Fox News | 17410 | 0.650598 | disagrees | https://www.allsides.com/news-source/fox-news-... | right-center | 26760 | 44170 |
| Washington Post | 21434 | 1.682022 | agrees | https://www.allsides.com/news-source/washingto... | left-center | 12743 | 34177 |
| New York Times - News | 12275 | 0.570002 | disagrees | https://www.allsides.com/news-source/new-york-... | left-center | 21535 | 33810 |
| HuffPost | 15056 | 0.834127 | somewhat disagrees | https://www.allsides.com/news-source/huffpost-... | left | 18050 | 33106 |
| Politico | 11047 | 0.598656 | disagrees | https://www.allsides.com/news-source/politico-... | left-center | 18453 | 29500 |
| Washington Times | 18934 | 2.017475 | strongly agrees | https://www.allsides.com/news-source/washingto... | right-center | 9385 | 28319 |
| NPR News | 15751 | 1.481889 | somewhat agrees | https://www.allsides.com/news-source/npr-media... | center | 10629 | 26380 |
| Wall Street Journal - News | 9872 | 0.627033 | disagrees | https://www.allsides.com/news-source/wall-stre... | center | 15744 | 25616 |
| Townhall | 7632 | 0.606967 | disagrees | https://www.allsides.com/news-source/townhall-... | right | 12574 | 20206 |
Visualizing the data
To make a bar plot we'll use Matplotlib with Seaborn's dark grid style:
As mentioned above, we have too many news outlets to plot comfortably, so just make a copy of the top 25 and place it in a new df2 variable:
| agree | agree_ratio | agreeance_text | allsides_page | bias | disagree | total_votes | |
|---|---|---|---|---|---|---|---|
| name | |||||||
| CNN (Web News) | 22907 | 0.970553 | somewhat disagrees | https://www.allsides.com/news-source/cnn-media... | left-center | 23602 | 46509 |
| Fox News | 17410 | 0.650598 | disagrees | https://www.allsides.com/news-source/fox-news-... | right-center | 26760 | 44170 |
| Washington Post | 21434 | 1.682022 | agrees | https://www.allsides.com/news-source/washingto... | left-center | 12743 | 34177 |
| New York Times - News | 12275 | 0.570002 | disagrees | https://www.allsides.com/news-source/new-york-... | left-center | 21535 | 33810 |
| HuffPost | 15056 | 0.834127 | somewhat disagrees | https://www.allsides.com/news-source/huffpost-... | left | 18050 | 33106 |
With the top 25 news sources by amount of feedback, let's create a stacked bar chart where the number of agrees are stacked on top of the number of disagrees. This makes the total height of the bar the total amount of feedback.
Below, we first create a figure and axes, plot the agree bars, plot the disagree bars on top of the agrees using bottom, then set various text features:
For a slightly more complex version, let's make a subplot for each bias and plot the respective news sources.
This time we'll make a new copy of the original DataFrame beforehand since we can plot more news outlets now.
Instead of making one axes, we'll create a new one for each bias to make six total subplots:
Hopefully the comments help with how these plots were created. We're just looping through each unique bias and adding a subplot to the figure.
When interpreting these plots keep in mind that the y-axis has different scales for each subplot. Overall it's a nice way to see which outlets have a lot of votes and where the most disagreement is. This is what makes scraping so much fun!
We have the tools to make some fairly complex web scrapers now, but there's still the issue with Javascript rendering. This is something that deserves its own article, but for now we can do quite a lot.
There's also some project organization that needs to occur when making this into a more easily runnable program. We need to pull it out of this notebook and code in command-line arguments if we plan to run it often for updates.
These sorts of things will be addressed later when we build more complex scrapers, but feel free to let me know in the comments of anything in particular you're interested in learning about.
Resources
Web Scraping with Python: Collecting More Data from the Modern Web — Book on Amazon
Jose Portilla's Data Science and ML Bootcamp — Course on Udemy
Easiest way to get started with Data Science. Covers Pandas, Matplotlib, Seaborn, Scikit-learn, and a lot of other useful topics.
Get updates in your inbox
Join over 7,500 data science learners.
Meet the Authors
Modern computers are equipped with processors that allow fast parallel computation at several levels: Vector or array operations, which allow to execute similar operations simultaneously on a bunch of data, and parallel computing, which allows to distribute data chunks on several CPU cores and process them in parallel. When working with large amounts of data, it is important to know how to exploit these features because this can reduce computation time drastically. Taking advantage of this usually requires some extra effort during implementation. With packages like NumPy and Python’s multiprocessing module the additional work is manageable and usually pays off when compared to the enormous waiting time that you may need when doing large-scale calculations inefficiently.
Dump the loops: Vectorization with NumPy
Many calculations require to repeatedly do the same operations with all items in one or several sequences, e.g. multiplying two vectors a = [1, 2, 3, 4, 5] and b = [6, 7, 8, 9, 10]. This is usually implemented with a loop (e.g. for or while loop) where each item is treated one by one, e.g. 1 * 6, then 2 * 7, etc. Modern computers have special registers for such operations that allow to operate on several items at once. This means that a part of the data, say 4 items each, is loaded and multiplied simultaneously. For the mentioned example where both vectors have a size of 5, this means that instead of 5 operations, only 2 are necessary (one with the first 4 elements and one with the last “left over” element). With 12 items to be multiplied on each side we had 3 operations instead of 12, with 40 we had 10 and so on.
In Python we can multiply two sequences with a list comprehension:
Python Web Data
This is fine for smaller data. However, it is not as efficient as vectorizing the multiplication with NumPy. When we put the data into NumPy arrays, we can write the multiplication as follows:
First of all, that’s much more compact than writing a list comprehension. Furthermore, it’s also much faster due to vectorization, as we can see when we multiply two arrays with 1,000,000 integers each. Let’s start with the unoptimized, pure Python implementation using random integers:
I’m using the %timeit “magic command” in an IPython session to measure the execution time which gives me about 63ms on my machine. Now to the vectorized version implemented with NumPy:
The execution time goes down to about 1.9ms, which means the calculations are more than 30x faster! At the same time, the extra effort for implementation was low and I would say that using the * operator for multiplying two NumPy arrays is more natural and concise than using a list comprehension or a loop.
A more practical example for vectorization
Let’s create a more practical example for vectorization to see how much can be achieved in an everyday task. One such task might be calculating the great circle distance (GCD) of two points on earth, which can be done with the haversine formula. The formula involves trigonometric operations, multiplications, square root, etc. so it might beneficial to use vectorization. Let’s have a look at a non-vectorized implementation in pure Python first:
The function takes a row of data, which is a tuple of 4 elements: the latitude and longitude of two points, a and b in degrees. It converts them to radians, then applies the calculations in the formula and returns the GCD in km. The function accepts the data as row because this way it is easier to apply it to our data, which will be a matrix where the four columns represent the origin latitude and longitude, and the destination latitude and longitude as in this example:
We can now apply the function to each row of the data, i.e. each row with its origin and destination coordinates will be passed to haversine. Again, this could be done with a list comprehension, but we can also use NumPy’s apply_along_axis, which is a little shorter to write. apply_along_axis takes three arguments: the function to apply, the axis on which this function is applied (for a 2D matrix 0 means column-wise and 1 means row-wise), and finally the data itself:
This correctly gives us about 930km for the distance Berlin–London (the first row of the data), 1609km for Berlin–Moscow (2nd row) and 2500km for Moscow–London (3rd row).
What we just did was taking each row of the input data, so four values per row, and then use these values for calculating the GCD. So each row is treated individually, just as with the initial example where two sequences were multiplied. In order to do the calculations more efficiently, we could vectorize them. Instead of thinking in row-wise calculations, we should think in column-wise calculations, where each column is a vector who’s values can be used simultaneously. So instead of converting a single origin’s latitude to radians with a_lat = math.radians(a_lat), we could take all origins’ latitudes, i.e. the whole column, and turn it into radians with a vectorized operation from NumPy like: a_lat = np.radians(a_lat). Note that a_lat is not a scalar (single) value anymore but contains all origins’ latitudes.
When we want to vectorize a function, we have to think about the layout of the data first and also about the calculations that are made. Not all calculations and algorithms can be vectorized efficiently. In our case, however, we are lucky since the input data can be split into columns as input vectors and all operations translate nicely into vectorized operations that are implemented in NumPy:
This vectorized version includes the same calculations as the previous version, but instead of a row with four values that represent single origin and destination coordinates, it takes vectors (NumPy arrays) of origin latitudes, origin longitudes, destination latitudes and destination longitudes. Most of the math functions have the same name in NumPy, so we can easily switch from the non-vectorized functions from Python’s math module to NumPy’s versions.
We now pass our function the columns of the data and it gives us the same result as before:
The expression coords[:, 0] says take all rows (: on the first axis) and the first column (0 on the second axis).
Now to the test: We can generate 10,000 random coordinates and measure the execution time of the “classic” and the vectorized implementation (see links on the bottom of the article for the whole scripts). This gives 53.8 ms and 2.9 ms, respectively, which means a speed up factor of about 18.
Similar principles can be applied to pandas DataFrames:
Now we can access the columns by name to pass them to the vectorized haversine function:
Comparing to a non-vectorized implementation (using DataFrame.apply), we get a speed up factor of more than 30 (174 ms vs. 4.8 ms).
All in all, the speed up that can be achieved with vectorization is immense. It might not be noticeable with small data and simple calculations. But when your data grows and calculations get more complex, you might end up waiting hours for results. Dividing this time by a factor of 30 is significant. At the same time, I think the extra effort in implementation work is quite low. The example showed how to vectorize the haversine formula, but you can apply the same principles to many other formulas and algorithms.
Divide and accelerate: Parallel computing with multiprocessing
By using vectorization, we exploit one important feature of modern processors (CPUs). However, we only use one CPU core, whereas nowadays desktop machines are usually equipped with at least 4 cores. We can exploit this with parallel processing, which I already briefly explained in connection with text analysis. The idea is that the data is split into equally sized chunks and then those chunks are distributed to the CPU cores so that each of them works on its own chunk and returns its calculation results. Those partial results are then combined to form the final result.
If you want to implement parallel processing on a single machine and distributing the workload is not too complex, a good way to start is to use Python’s multiprocessing module and its Pool class. We will use its method apply_async to distribute the work across several “worker processes”. All of these processes run the same function (i.e. the same calculations) but with different parts (“chunks”) of the data.
At first we will define a function that is called by each worker process. A chunk of data is passed to this function and we calculate the GCD for this chunk. Finally, we set the index of the result data to be the same as the index in the input chunk. This is necessary in order to combine the partial results from the individual processes later.
Now we can take care about distributing the work. In parallel processing, this is often one of the most difficult tasks, because you want to make sure that each worker process takes about the same time to finish the calculations. You can only create the final results once you get all the partial results from the worker processes. Hence if one process takes much more time to compute than the others, you’ll have to wait for it to finish while the other processes are idle.
In our case, we can just distribute the data evenly across the worker processes, because each row of data will take about the same time to compute. If we had, for example, text strings of different lengths in the data rows, this distribution scheme would not be efficient.
We’re still working with a dataframe of coordinates, called df_coords like in the previous examples. At first we determine the number of worker processes that we want to start. We’ll use all CPU cores in our machine:
Next we determine the size of each chunk by integer division:
Of course, this will often result in a remainder, e.g. when we have 13 rows of data and 4 processes, then chunksize will be 3 but we’ll have 1 row as remainder. We will handle this now as follows: A list proc_chunks will contain a data chunk for each worker process. For each process we define a start row number chunkstart and an end row number chunkend. We’ll use these row numbers for slicing the dataframe. The last process will always get everything that’s left. Please note that the row numbers start with 0 and end with “num. rows – 1”. So given the former example, we’d end up with the following distribution of data chunks for the individual worker processes:
| process | rows in chunk |
|---|---|
| process #1 | 0, 1, 2 |
| process #2 | 3, 4, 5 |
| process #3 | 6, 7, 8 |
| process #4 | 9, 10, 11, 12 |
The distribution of the remainder is not optimal but we’ll leave it like this for the sake of simplicity. We can implement this as follows:
It’s always good to add an assertion to make sure all data ended up in the chunks (i.e. the sum of chunk lengths match):
We can now start a pool of parallel worker processes, pass each process its chunk of data and let it run the process_chunk function with it. It’s important to use apply_async for this, because this will distribute the data and start the processes simultaneously (“non-blocking”) without waiting for individual processes to finish. The results should be saved in a list proc_results that will contain the partial result of each worker process. Fetching those results is then a “blocking” task, i.e. we wait for the processes to finish the calculations by calling get() on each result object:
When all worker processes are finished with their calculations, their partial results will be saved in result_chunks. We can use concat to concatenate those results to a single dataframe and join it with the input data so that the final result will contain the coordinates and their respective distances:
Running this while investigating the operating system’s activity with a tool like htop will show that all processors will be busy during execution instead of just one. So this should speed up our calculations even more, right? Unfortunately not necessarily! For example, with 1,000,000 random coordinate pairs my 4-core machine took about 50% longer for calculating the results in parallel than with single processor execution. What happened? The vectorized code already runs quite fast, even for large datasets. Splitting the data into chunks, starting the worker processes, distributing the data and then collecting and combining the results again introduces a lot of extra work (“overhead”). This extra work unfortunately does not pay off in this scenario, because the actual processing time for each chunk is quite low, compared to the time spend for the parallelization overhead.
Still, there are many scenarios where parallelization does pay off despite the overhead. This is usually the case when the processing time for the data is high as compare to the parallelization overhead. If you have, for example, algorithms or formulas that cannot be vectorized, then parallelization will usually pay off. Let’s say for example, that we wanted to generate a hash for each observation in our data (i.e. each row – not each value). If you have a dataframe, you could do so with df.apply(lambda row: hash(tuple(row)), axis=1).* Running this in parallel gives a speed up factor of ~3 on my 4-core machine (again, the theoretical speed up of 4 is not reached because of overhead).
So parallelization can also be very helpful when it comes to reducing the calculation time. This is especially helpful when you have access to cluster machines that often have dozens of CPUs. On the other hand, you should consider wisely if parallelization will really help, because it usually takes more effort to implement it and can cause more headaches due to asynchronous execution. Furthermore, it might be faster, but use more memory so this can be a trade-off decision.
There are lots of Python packages for parallel and distributed computing, and you should consider using them when Python’s default multiprocessing module does not fit your needs:
- joblib provides an easier to use wrapper interface to multiprocessing and shared memory
- dask is a complex framework for parallel and distributed computing
Beautifulsoup To Dataframe
I uploaded the full scripts as gists:
Pandas Web Scraping Tools
- unoptimized.py contains non-vectorized haversine and hashing
- vectorized.py contains vectorized haversine calculation
- parallelized.py contains parallel execution of vectorized haversine calculation and parallel hashing
* Of course this is a made up example since you could also vectorize the hashing function. But this is only a short placeholder for algorithms that cannot (easily) be vectorized, and should prove the point that parallelization can also reduce execution time drastically.
